Resources
This page contains links to code or other useful materials we developed.
Code
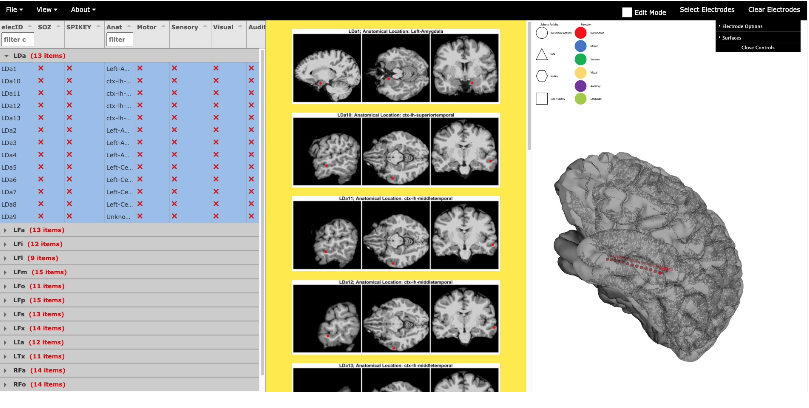
Viewing and intuitive interaction with multimodal data from intracranial patients is challenging. We developed a browser based viewer that: • Does not require installation of additional software • Has dynamic 3D rendering for easy adjustments of views • Runs locally (no need for internet, no need to upload sensitive data to a server) • Works on every OS • Does not require technical knowledge
Accurate intracranial electrode localization and visualization is a pre-requisite for all iEEG research. iElvis is a toolbox that helps with both those aspects. Groppe, DM. et al. J Neurosci Meth 281, 40–48 (2017).

Neurodata without Borders (NWB) is a newly developed standard to save neurophysiology data. We developed Python code to import our iEEG data, other neurophysiological signals, and meta-data from command line or via a GUI. We tried to make it as useable for other labs as possible. Try it out and please let us know if you have suggestions! Thank you to the Kavli Foundation’s Seed Grant to Stephan, which helped developing this!
Our iEEG preprocessing and analysis code. (We’re currently cleaning it up for you. Stay tuned.)
Data
We are committed to sharing our data in NWB format and are in the process of converting our old and new data. We will list all the data we share here. Stay tuned.
- Code and data for the paper Intracranial electroencephalography reveals effector-independent evidence accumulation dynamics in multiple human brain regions.. Gherman et al. Nat. Hum. Behav. 8, 758–770 (2024). (stay tuned for an NWB version)
- ...in process. For now check our the links to code and data on our publications. </ul>